Absolute Bacterial Abundance
The sequencing solutions we use to profile the microbiome of a sample provide information on the relative abundance of the microorganisms in each sample. Relative abundance tells us how many percentages of the microbiome are made up of a specific organism, e.g. if E. coli makes up 1% or 10% of the total amount of bacteria detected in a sample. This allows us to evaluate if the relative abundance of each organism changes or is associated with a variable of interest such as diet, medication, pollution, pesticide usage, etc. If the medication kills other bacteria but not E. coli or gives E. coli a special advantage following medical treatment, we will see an increase in the relative abundance of E. coli in the treated subjects as compared to the untreated subjects.
When insight is necessary
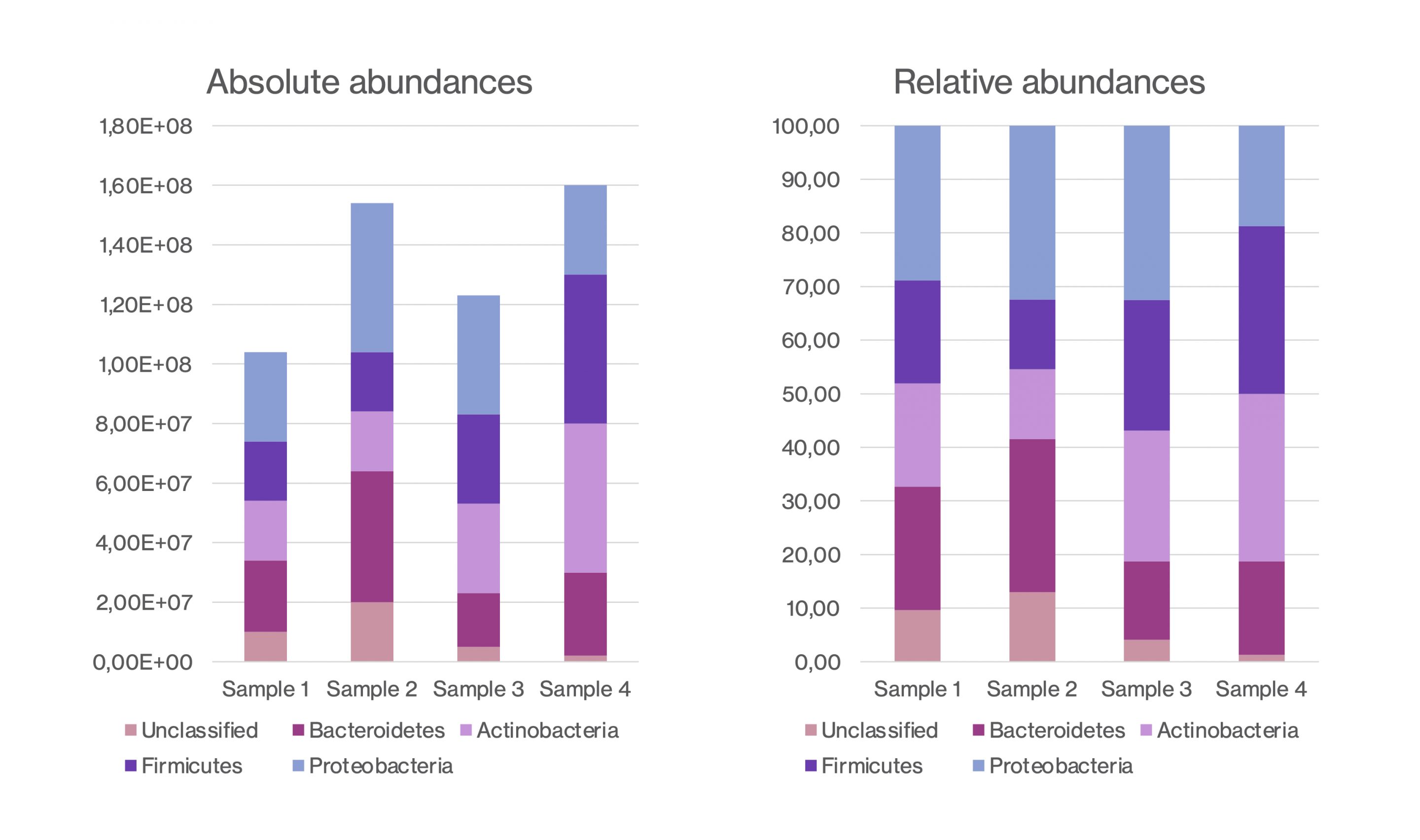
But sometimes insight into the absolute abundance of organisms in a sample is necessary to fully understand how the microbiome is affected. Such a situation may arise if a treatment does not change the relative abundance of the microorganisms but changes the absolute bacterial abundance. Similarly, if the abundance of one bacterium increases it will decrease the relative abundance of other bacteria, but perhaps not their actual amount (see figure below). But based on relative abundance data it is difficult to detect if a bacterium changed its absolute abundance. Without absolute quantification, we simply observe that their relative abundance changed.
Statistical solutions have been developed to facilitate the identification of such changes from relative abundance data, however, these methods have limitations, and it can therefore be an advantage to use methods that allow for absolute quantification. This is specifically the case in studies with small sample sizes.
The question of when you have a small sample size is always complex, but we have a few “rules of thumb” that say that for comparisons of different groups e.g. cases versus controls, less than 20 samples per group is a small sample size. However, if you have a very noisy dataset with many confounders, or expect a low effect size, then you need to increase this number. For evaluation of quantitative traits, less than 50 samples are considered small, but again you need to consider the noise, the effect size, and the method used (how many variables do you wish to analyze).
We are always happy to help you design your microbiome study, so we increase the chances of robust results.
Spike-in of bacteria allows absolute quantification
A leading solution to address the problem of relative abundance is to add a spike-in of bacteria of known concentration to the sample. When the bacteria are rare and not found in the environment being investigated, this allows for statistical re-calculation of the data to obtain both relative and absolute abundance tables (absolute quantification). The method is still at an early stage and has its limitations. For example, when the sequencing method used is amplicon sequencing (16S or ITS), absolute quantification is only available for bacterial profiling, as the spike-in organisms are bacteria. When using shotgun sequencing, all organisms are profiled in the same sequencing run and we can use the spike-in to quantify all detected organisms.
We have found that including absolute quantification can provide valuable information, especially if a shift in absolute abundance is expected e.g. in evacuations of antibiotic treatment effects, or if the sample size is low as discussed above.
Figure of the difference between relative abundance and absolute abundance profiles.
While “reference frames”-based statistical solutions can compensate for potential shortcommings of relative abduance data for some questions, in some studies, in others such as studies with small samples sizes or in projects profilin less diverse microbiome communities; spike-in solutions are necessary.


Solving your microbiome problems
We have more than +7 years of experience in analyzing microbiome data and structuring microbiome projects. Our team has worked on more than 75 microbiome projects spanning research, universities and commercial industries.
Biomcare offers both microbiome services for small discovery projects, as well as large custom-designed microbiome projects.
Contact us today and get a quote. We are standing by to service you.